Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP - ScienceDirect

Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP: Cell

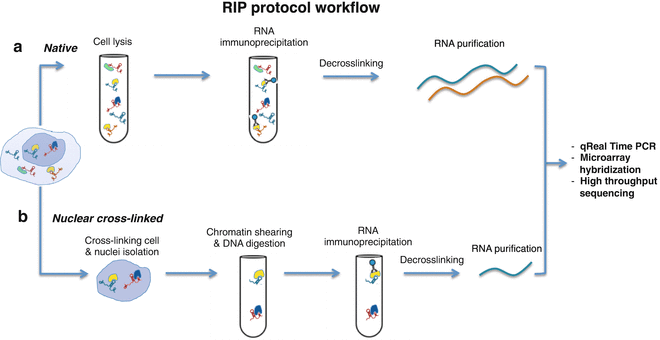
RNA structure, binding, and coordination in Arabidopsis - Foley - 2017 - WIREs RNA - Wiley Online Library
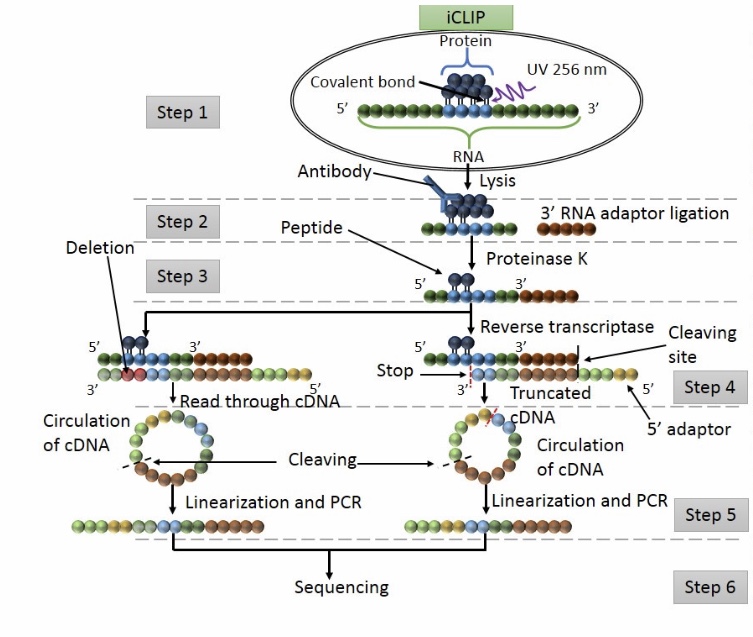
Protein – RNA interactions: Analysis of iCLIP-seq data FU Berlin Seminar RNA Bioinformatics, WS 14/15 Sabrina Krakau

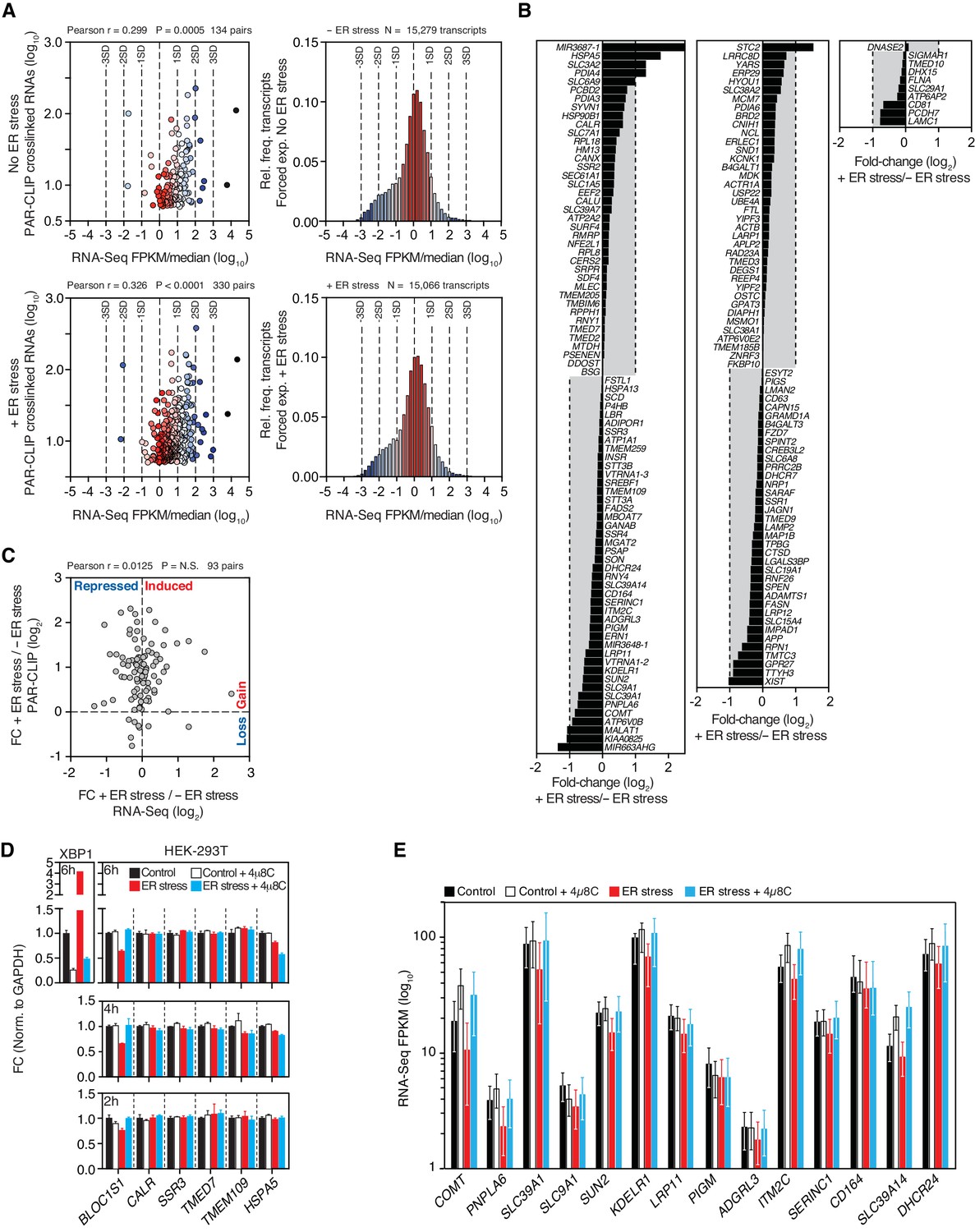
PAR-CLIP and streamlined small RNA cDNA library preparation protocol for the identification of RNA binding protein target sites - ScienceDirect

Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins - ScienceDirect
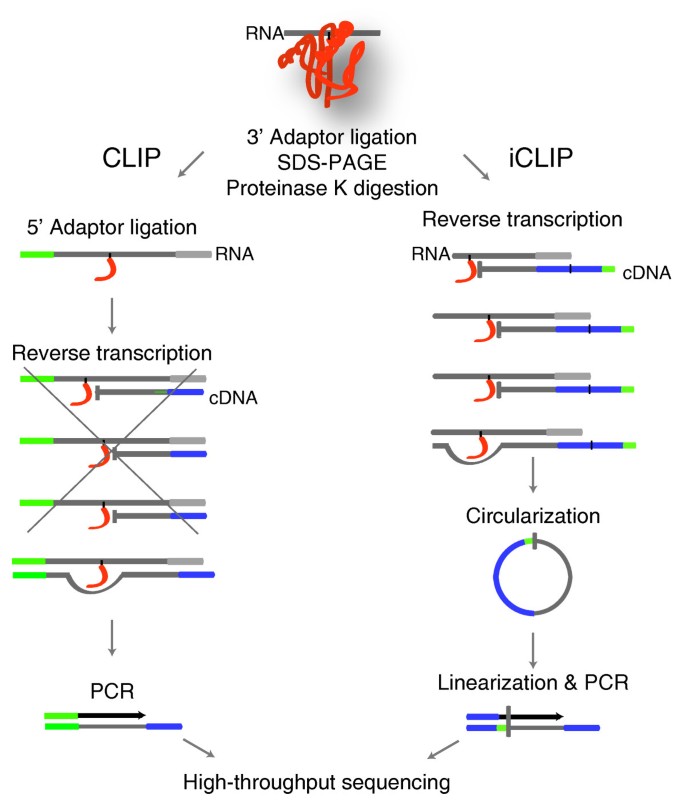
Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions | Genome Biology | Full Text


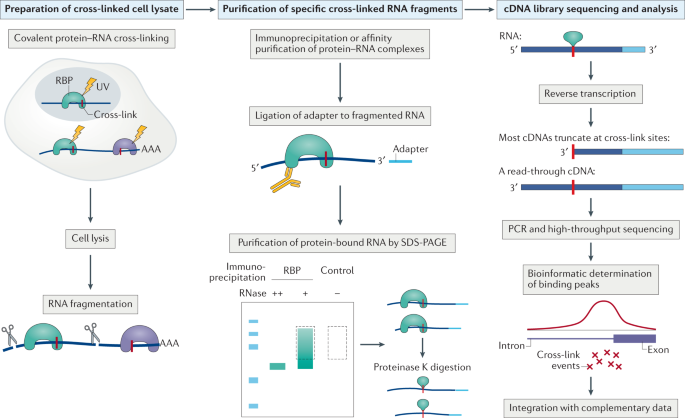

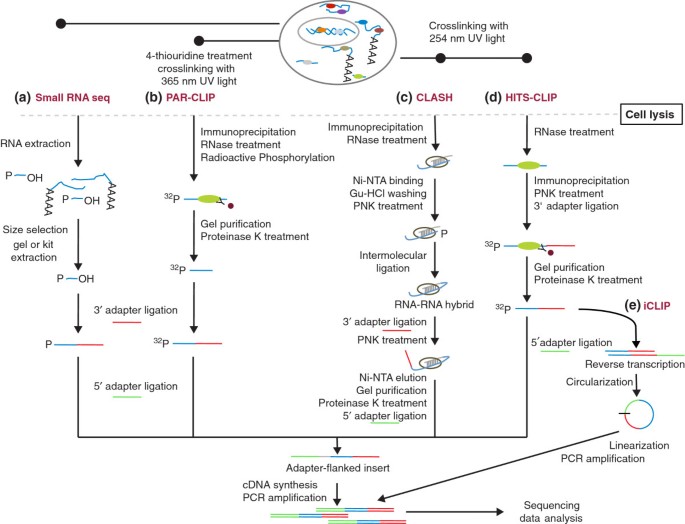
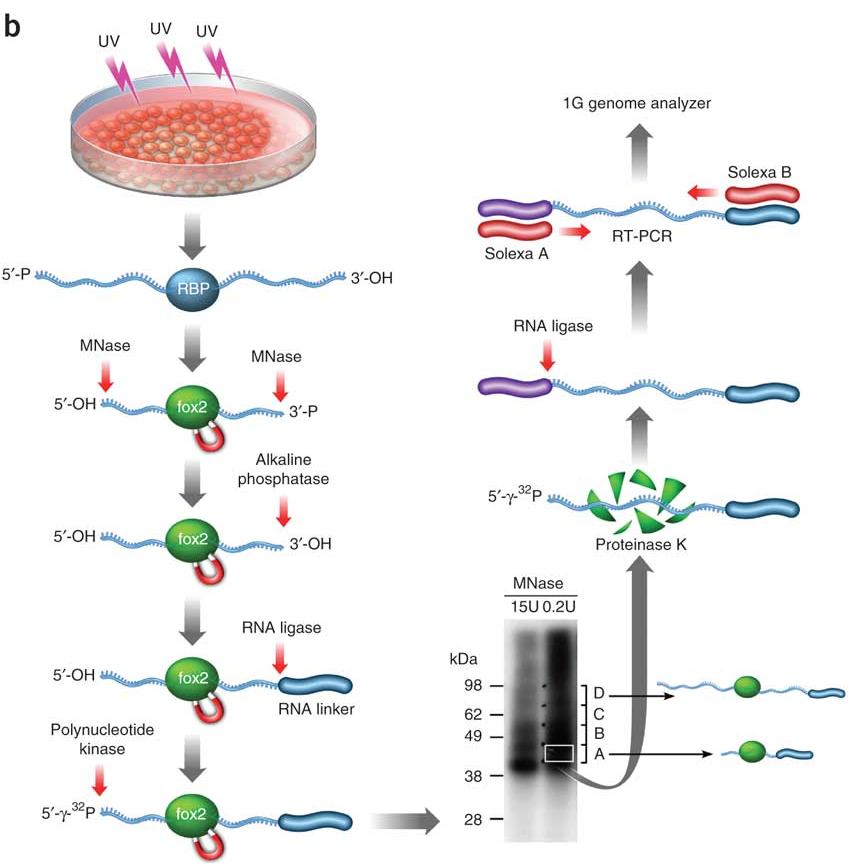

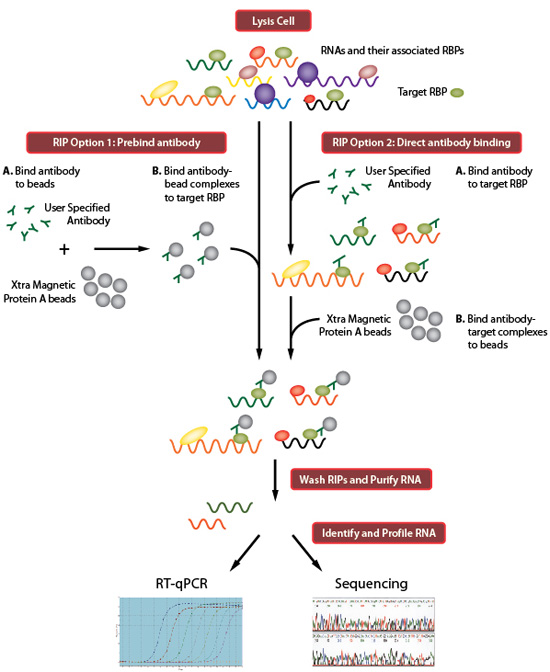
![PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/e504d75c2c48b19613a796fb54083b71e3781289/2-Figure1-1.png)